European Journal of Human Genetics (2011) 19, 216–223; doi:10.1038/ejhg.2010.153; published online 8 September 2010
A Central Asian origin of the Hazaras?
In the heartland of Eurasia: the multilocus genetic landscape of Central Asian populations
Begoña Martínez-Cruz1,7,10, Renaud Vitalis1,8,10, Laure Ségurel1,9, Frédéric Austerlitz2, Myriam Georges1, Sylvain Théry1, Lluis Quintana-Murci3, Tatyana Hegay4, Almaz Aldashev5, Firuza Nasyrova6 and Evelyne Heyer1
1Muséum National d’Histoire Naturelle – Centre National de la Recherche Scientifique-Université Paris 7, UMR 7206, ‘Éco-Anthropologie et Ethnobiologie’, Paris, France
2Laboratoire Écologie, Systématique et Évolution, Université Paris Sud, CNRS UMR 8079, Orsay, France
3Human Evolutionary Genetics, Institut Pasteur, CNRS URA3012, Paris, France
4Uzbek Academy of Sciences, Institute of Immunology, Tashkent, Uzbekistan
5National Center of Cardiology and Internal Medicine, Bishkek, Kyrgyzstan
6Tajik Academy of Sciences, Institute of Plant Physiology and Genetics, Dushanbe, Tajikistan
Correspondence: Professor E Heyer, Muséum National d’Histoire Naturelle – Centre National de la Recherche Scientifique, Université Paris 7, UMR 7206, ‘Éco-Anthropologie et Ethnobiologie’, CP 139, 57 rue Cuvier, 75231 Paris Cedex 05, France. Tel: +33 (0)1 40 79 81 58 ; Fax: +33 (0)1 40 79 32 31; E-mail: heyer@mnhn.fr
7Current address: Evolutionary Biology Institute, Pompeu Fabra University – CSIC – PRBB, Barcelona, Spain.
8Current address: Centre National de la Recherche Scientifique – Institut National de la Recherche Agronomique, UMR CBGP (INRA – IRD – CIRAD – Montpellier SupAgro), Campus International de Baillarguet, Montferrier-sur-Lez, France.
9Current address: Department of Human Genetics, University of Chicago, Chicago, IL, USA.
10These authors contributed equally to this work.
Received 25 January 2010; Revised 21 July 2010; Accepted 5 August 2010; Published online 8 September 2010.
Abstract
Located in the Eurasian heartland, Central Asia has played a major role in both the early spread of modern humans out of Africa and the more recent settlements of differentiated populations across Eurasia. A detailed knowledge of the peopling in this vast region would therefore greatly improve our understanding of range expansions, colonizations and recurrent migrations, including the impact of the historical expansion of eastern nomadic groups that occurred in Central Asia. However, despite its presumable importance, little is known about the level and the distribution of genetic variation in this region. We genotyped 26 Indo-Iranian- and Turkic-speaking populations, belonging to six different ethnic groups, at 27 autosomal microsatellite loci. The analysis of genetic variation reveals that Central Asian diversity is mainly shaped by linguistic affiliation, with Turkic-speaking populations forming a cluster more closely related to East-Asian populations and Indo-Iranian speakers forming a cluster closer to Western Eurasians. The scattered position of Uzbeks across Turkic- and Indo-Iranian-speaking populations may reflect their origins from the union of different tribes. We propose that the complex genetic landscape of Central Asian populations results from the movements of eastern, Turkic-speaking groups during historical times, into a long-lasting group of settled populations, which may be represented nowadays by Tajiks and Turkmen. Contrary to what is generally thought, our results suggest that the recurrent expansions of eastern nomadic groups did not result in the complete replacement of local populations, but rather into partial admixture.
Keywords:
admixture; Central Asia; ethnic groups; genetic diversity; microsatellites; population genetics
INTRODUCTION
The evolutionary history of modern humans has been characterized by range expansions, colonizations and recurrent migrations over the last 100?000 years.1Some regions of the world that have served as natural corridors between landmasses are of particular importance in the history of human migrations. Central Asia is probably at the crossroads of such migration routes.1, 2 Located in the Eurasian heartland, it encompasses a vast territory, limited to the east by the Pamir and Tien-Shan mountains, to the west by the Caspian Sea, to the north by the Russian taiga and to the south by the Iranian deserts and Afghan mountains. The role of Central Asia in both the early spread of modern humans out of Africa and the more recent settlement of differentiated populations3 is not precisely known.4, 5, 6 For example, it remains unclear as to whether this region harbored a Paleolithic ‘maturation phase’ of modern humans before giving rise to waves of migration, resulting in colonization of the Eurasian continent6 or whether it has served as a meeting place for previously differentiated Asian and European populations following their initial expansions.3, 7
Central Asia entered the historical records about 1300 bc, when Aryan tribes invaded the Iranian territory from what is nowadays Turkmenistan and established the Persian Empire in the seventh century bc.8 A branch of those, the Scythians, described in ancient Chinese texts and in Herodotus’ Histories, as having European morphological traits and speaking Indo-Iranian languages, expanded north into the steppes. Thereafter, Central Asia was faced with multiple waves of Turkic migrations, although it is difficult to know precisely when these westward expansions began. Between the second and the first century bc, Huns brought the East-Asian anthropological phenotype to Central Asia.8 At the same period, the Chinese established a trade route (the Silk Road), which connected the Mediterranean Basin and Eastern Asia for more than 16 centuries. In the thirteenth century ad the Turco-Mongol Empire lead by Genghis Khan became the largest of all time, from Mongolia to the Black Sea. All these movements of populations resulted in a considerable ethnic diversity in Central Asia, with Indo-Iranian speakers living as sedentary agriculturalists and Turkic speakers mainly living as traditionally nomadic herders.
Taken together with the ancient peopling of Central Asia, this intricate demographic history shaped patterns of genetic variability in a complex manner. Most previous studies, based on classical markers,1 mitochondrial DNA (mtDNA)3,9, 10, 11, 12, 13 or the non-recombining portion of the Y-chromosome (NRY),6, 14,15, 16 have shown that genetic diversity in Central Asia is among the highest in Eurasia.3, 6, 15 NRY studies suggest an early settlement of Central Asia by modern humans, followed by subsequent colonization waves in Eurasia,6 whereas some mtDNA studies point to an admixed origin from previously differentiated Eastern and Western Eurasian populations.11 Furthermore, a recent analysis of mtDNA data suggests east-to-west expansions waves across Eurasia.14 However, inferring more accurately the impact of population movements, including the expansion of eastern nomadic groups, requires additional, fast-evolving molecular markers. Here we report on the first multilocus autosomal genetic survey of Central Asian populations. Twenty-six populations from six ethnic groups were genotyped at 27 autosomal unlinked microsatellite markers. We aimed to shed light on the genetic origins of Central Asian populations, and to investigate how the recurrent westward expansions of eastern nomadic groups during historical times have shaped the Central Asian genetic landscape.
MATERIALS AND METHODS
DNA samples
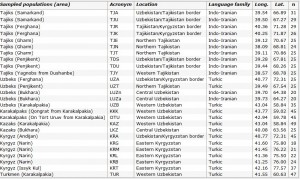
We sampled 767 men belonging to 26 populations from western Uzbekistan to eastern Kyrgyzstan (Table 1 and Figure 1) representative of the ethnological diversity in Central Asia: Tajiks, which are Indo-Iranian speakers (a branch of the Indo-European language family), and Kazakhs, Turkmen, Karakalpaks, Kyrgyz and Uzbeks, which are Turkic speakers (a branch of the Altaic language family). In two Uzbek populations from the Bukhara area (LUZa and LUZn), an extensive linguistic survey showed that individuals were bilingual, speaking both Tajik and Uzbek. As their home language was Tajik (an Indo-Iranian language), we further classified these two populations into the Indo-Iranian group for subsequent analyses. We collected individuals unrelated for at least two generations back in time. All individuals gave informed consent for their participation in this study. Total genomic DNA was isolated from blood samples by a standard salting out procedure17 followed by a phenol–chloroform extraction.18

Genotyping
We selected 27 microsatellite markers19 from the set of 377 markers used in the worldwide study by Rosenberg et al.20 The choice and description of markers, PCR and electrophoresis conditions are given in Ségurel et al.19 We further genotyped 20 individuals from the HGDP-CEPH Human Genome Diversity Cell Line Panel20,21, 22 at the 27 microsatellite loci, in order to standardize the original Central Asian data presented here with the worldwide HGDP-CEPH data.
Data analyses
Genetic diversity
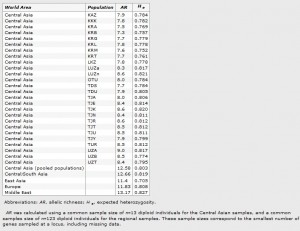
In each population and for each locus, we calculated the allelic richness (AR) using the rarefaction method proposed by El Mousadik et al23 with the software package FSTAT.24 Unbiased estimates of expected heterozygosity (He)25 were computed in each population for each locus with GENETIX.26 Both ARand He estimates were averaged over the loci in each population. We tested heterogeneity in both AR and He among populations using the Kruskal–Wallis test, with locus-specific estimates taken as replicate observations. Locus-specific ARand expected heterozygosity were also estimated for populations pooled into Indo-Iranian- and Turkic-speaking groups, and averaged over loci within groups. We tested between-group differences in both AR and He using the Wilcoxon’s signed-rank test, with locus-specific estimates taken as replicate observations. We further estimated AR and He for each locus over the pooled data from Central Asia and over the pooled data for Central/South Asia, East Asia, Europe and the Middle East from the HGDP-CEPH Panel, and calculated the averages over loci within groups. We tested heterogeneity in both AR and He across the five groups of Eurasian populations using the Kruskal–Wallis test, taking locus-specific estimates as replicate observations. When significant differences among groups were found, we ran the Tukey’s range test to find which group statistics were significantly different from one another. All statistical analyses were performed with the software package – JMP5.1 (SAS Institute Inc.).27
Genetic structure
Population differentiation (FST) was calculated overall and between pairs of Central Asian populations with GENEPOP 4.0.28 Exact tests of differentiation were performed with FSTAT,24 adjusting P-values with Bonferroni correction for multiple tests. We performed a correspondence analysis (CA) based on tables of allele counts using GENETIX.26 The population structure was also inferred by means of a hierarchical analysis of molecular variance (AMOVA),29 with populations pooled into ethnic or linguistic groups. For ethnic grouping, populations were pooled as Tajiks (TJA, TDS, TJT, TJK, TJR, TJN, TDU, TJE, TJY and TJU), Karakalpaks (KKK and OTU), Kazakhs (KAZ and LKZ), Kyrgyz (KRA, KRG, KRL, KRB, KRT and KRM), Uzbeks (UZA, UZB, LUZa, LUZn and UZT) and Turkmen (TUR). For linguistic grouping, populations were pooled as Indo-Iranian speakers (Tajiks and the two Uzbek populations LUZa and LUZn) and Turkic speakers (all other populations). These analyses were performed with ARLEQUIN 3.11.30Isolation-by-distance (IBD) was tested with GENEPOP 4.0.28 We used PATHMATRIX31 to compute the matrix of effective geographical distances, based on a least-cost path algorithm. The least-cost distances, which account for the cost of the movement through the slopes in the landscape, were calculated from the digital elevation model GTOPO30 of the Earth Resources Observation and Science Center.
Clustering analyses
We performed a clustering analysis with STRUCTURE32 on the Central Asian populations together with all the Eurasian and African populations from the HGDP-CEPH Panel H952 corrected data set.33, 34 We used the latest version of STRUCTURE35 (version 2.3), which allows structure to be detected at lower levels of divergence than the original model. Each Markov chain was run for 106 steps, after a 105-step burn-in period. In each case, the results were checked to ensure consistency over 40 independent runs. Potential distinct modes among the 40 runs were identified using the Greedy algorithm implemented in CLUMP.36 We varied the hypothetical number of clusters (K) from 1 to 8 for all analyses. All chains were run using the F model for correlations of allele frequencies across clusters.37
Admixture analyses
The Central Asian genetic pool may be more than just the result of admixture from Eurasian populations, but we were nonetheless interested in investigating the potential origins of Central Asian populations among all Eurasian populations. We used LEADMIX38 to calculate maximum likelihood estimates (MLE) of the admixture proportions for each Central Asian population. We ran the program independently for each of them, considering four putative parental groups from the HGDP-CEPH Panel: Central/South Asia, East Asia, Europe and Middle East. For the Central/South Asian group, we chose a pool of Balochi (n=25) and Makrani (n=25) individuals, both populations being non-significantly differentiated (FST=?0.002; exact test P=0.34). We chose the Han Chinese (n=44) for the East-Asian parental group, and we further considered a pool of French (n=28), Bergamo (n=13) and Tuscan (n=21) individuals for the European group, these three populations being non-significantly differentiated (FST <?0.006;P>0.42). Last, we chose the Palestinians (n=46) for the Middle Eastern group.39
RESULTS
Genetic diversity
Average AR and expected heterozygosity for each of the 26 Central Asian populations and across regions are given in Table 2. We found a significant difference in AR (Kruskal–Wallis test, ?2=105.29, d.f.=25, P<0.0001) and in expected heterozygosity (Kruskal–Wallis test, ?2=67.98, d.f.=25, P<0.0001) among populations. We found no significant difference in AR between Indo-Iranian (AR=13.8) and Turkic speakers (AR=13.7, Wilcoxon signed-rank test, Z=?0.69,P=0.49), although the expected heterozygosity was significantly higher in Indo-Iranian as compared with Turkic speakers (He=0.818 and 0.787, respectively, Wilcoxon signed-rank test, Z=?4.55, P<0.0001). We found a significant difference in AR across Central Asia, Europe, Central/South Asia, Middle East and East Asia (Kruskal–Wallis test, K=36.46, d.f.=4, P<0.0001), as well as in expected heterozygosity (Kruskal–Wallis test, K=52.94, d.f.=4, P<0.0001). Yet, these differences were rather owing to a lower heterozygosity in East Asia and also slightly higher AR in Middle East (Tukey’s test, P<0.0001 for both AR and He). Central Asia therefore showed neither higher nor lower diversity than the rest of Eurasia.